Introduction
- Fragment-based drug design (FBDD) uses molecular fragments to identify potential drug candidates, formulated as a combinatorial optimization problem. Finding the optimal solution in such a vast search space is challenging, particularly for generating a Pareto front of solutions, which classical samplers struggle to achieve.
- To address these challenges, we propose a novel method that leverages quantum techniques, offering more efficient navigation of the solution space and a broader set of promising candidates for evaluation.
Method
- Generate fragment library
- Translate constraints to QAOA
- Prepare Ansatz
- Sample library via QAOA
- Apply subgraph mining to reduce quantum errors
Conclusions
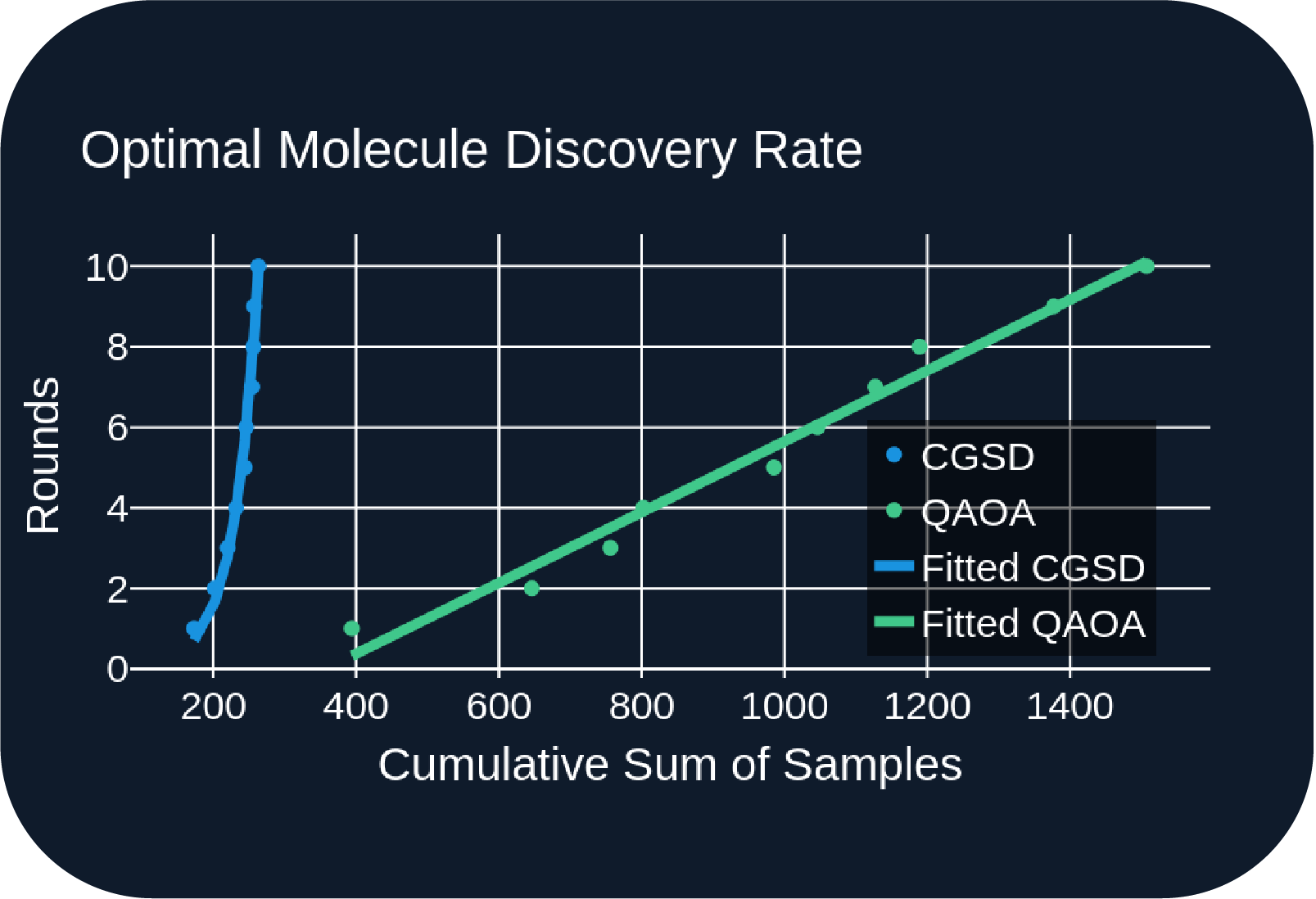
- We compared our hybrid QAOA sampler to the classical greedy steepest descent (CGSD) sampler:
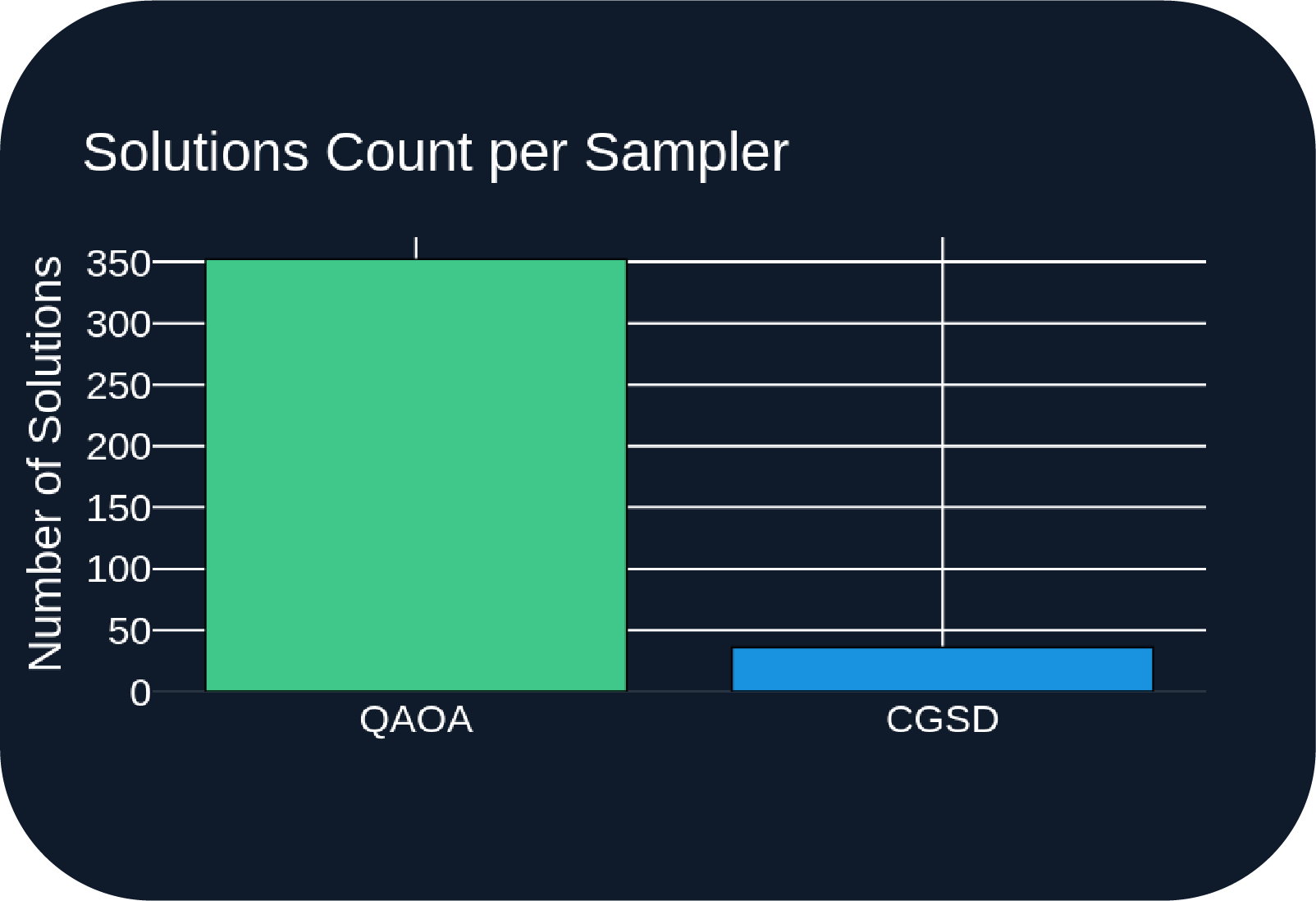
- Figure A: QAOA sampler returns over 5x more samples on average.
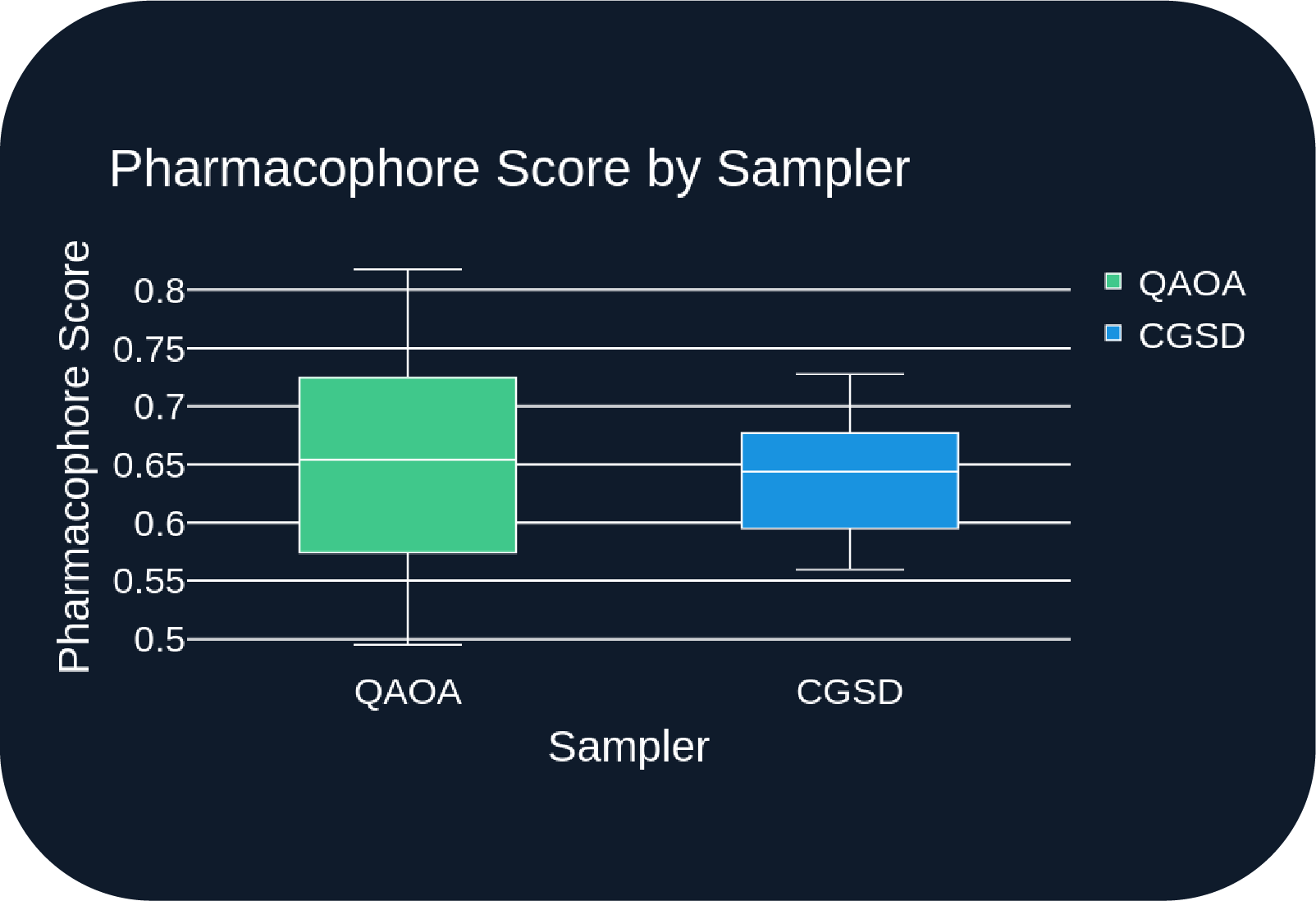
- Figure B: QAOA yields a broader range of pharmacophore scores, including higher scores than CGSD.
- Figure C: After 10 rounds, QAOA steadily discovers unique optimal molecules, while CGSD struggles to find new ones.
- This work is a part of a DARPA IMPAQT contract. More details here.